Discussion
Pegasystems Inc.
NL
Last activity: 19 Aug 2022 6:19 EDT
Creating PMML from Python, R and Pega
PMML is an XML based exchange format for analytic models supported by Pega. You can import models created outside of Pega by exporting them to PMML then importing the PMML files into Prediction Studio.
In this post we show minimalistic examples of creating PMML from Python and R and how to use these models in Pega.
Creating a PMML file from Python scikit-learn
Python scikit-learn is a popular machine learning toolkit for Python built on the also very popular NumPy and SciPy packages. With a few lines of code, we create a random forest model for customer churn. There are some preprocessing steps in the code that will also become part of the PMML file.
import pandas
import numpy
from sklearn2pmml.pipeline import PMMLPipeline
from sklearn.ensemble import RandomForestClassifier
from sklearn_pandas import DataFrameMapper
from sklearn.impute import SimpleImputer
churndata = pandas.read_csv("../cdh-datascientist-tools/dmsample/data/ChurnDMSample2.csv")
# Only use a subset of the data for modeling
devset = churndata[["Age", "AvgCallsOut"]]
# Map the multiple values of the Churn field
y = churndata["Churn"].map(lambda x: ("Churned", "Loyal")[x.startswith("N")])
# Create a preprocessor to replace missing values with median
pp = DataFrameMapper(
[(["Age", "AvgCallsOut"],
[SimpleImputer(missing_values=numpy.nan, strategy='median')])])
# Create a random forest classifier
churn_classifier = RandomForestClassifier(n_estimators=20)
# Create a PMML pipeline including some preprocessing
pipeline = PMMLPipeline([
("preprocessing", pp),
('churn_classifier', churn_classifier)])
# Fit the model
pipeline.fit(devset, y)
We use a dataset from DMSample, the OOTB sample application for Decisioning that ships with Pega. The dataset is available from the Open Source CDH utilities. To obtain it, clone the project from GitHub: https://github.com/pegasystems/cdh-datascientist-tools, or just download the CSV from the web interface directly.
The model can now be exported to PMML through the sklearn2pmml package developed by openscoring.io with a single line of code:
from sklearn2pmml import sklearn2pmml
sklearn2pmml(pipeline, "churn_sklearn.pmml", with_repr = True)
This will create a PMML file that you can now import into Pega.
The missing value imputation is put in the PMML file through properties of the MiningSchema. Other types of preprocessing may find their way into the TransformationDictionary section of the PMML file.

The pipeline approach makes it easy to include pre-processing steps into the PMML file, so you don't have to (try to) replicate the Python pre-processing steps in Pega but instead include them with the model itself. Please refer to sklearn2pmml documentation for more details.
The OOTB Churn model in DMSample was built with Pega's own modeling tool, and this too includes ways to create derived ("virtual") fields that automatically become part of the model representation.
Creating a PMML file from R
In a very similar way, we can create a PMML file from R. We use the same dataset and again a simple Random Forest classifier that predicts customer churn from just age and aggregated call data.
library(caret)
library(randomForest)
library(r2pmml)
churndata <- read.csv("../cdh-datascientist-tools/dmsample/data/ChurnDMSample2.csv", stringsAsFactors = F)
# Only use a subset of the data for modeling
devset <- churndata[, c("Age","AvgCallsOut")]
# Map Churn field (Y,yes,N,no) to two outcomes
y <- ifelse(startsWith(churndata$Churn,"N"),"Loyal","Churned")
# Create a preprocessor to replace missing values with the median
pp <- preProcess(devset, method = c("medianImpute"))
# Use the preprocessor to transform the dataset
devset.xformed = predict(pp, newdata = devset)
# Train a random forest with the Churn data
rf <- randomForest( devset.xformed, factor(y), ntree=20)
# Export the model to PMML, including preprocessing steps
r2pmml(rf, "churn_r.pmml", preProcess = pp)
Like the Python example, we do missing value imputation and include that in the PMML file. The r2pmml library supports this via the preProcess function from the caret library - which makes it a very powerful tandem.
The generated PMML looks slightly different, as here the PMML library is using the TransformationDictionary section of the PMML file. The result is the same.
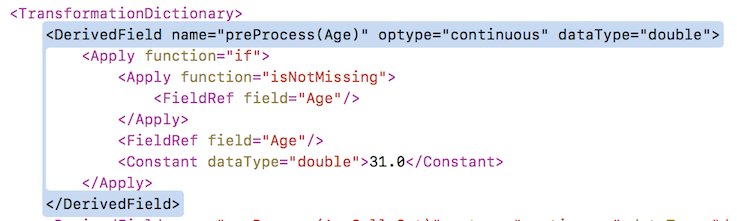
The r2pmml library is freely available and from the same authors as the sklearn library. It is a much better alternative than the older pmml library. For more info see the examples in the r2pmml documentation.
Exporting ADM Models as PMML
There is experimental support to export ADM models as PMML. A single ADM rule (or "configuration") can be exported to a PMML file. This PMML file is then an ensemble of Scorecards with each Scorecard representing an individual model instance.
The export can work off the Pega database or from an export of the tables in the ADM data mart. For generalizability, the code below works from such an export. To create the export
- Initialize DMSample so there are Adaptive models in the system, then
- Create a Pega Dataset (of type DB) on the classes Data-Decision-ADM-ModelSnapshot and Data-Decision-ADM-PredictorBinningSnapshot (future release may contain such datasets OOTB), then
- Run Export and download the resulting files.
library(XML)
library(cdhtools)
models <- readDSExport("Data-Decision-ADM-ModelSnapshot_All",
srcFolder="~/Downloads", tmpFolder="tmp")
predictors <- readDSExport("Data-Decision-ADM-PredictorBinningSnapshot_All",
srcFolder="~/Downloads", tmpFolder="tmp")
# Create a single PMML model to represent all
# the instances of the ADM SalesModel rule
adm2pmml(models, predictors, ruleNameFilter = "SalesModel")
The "cdhtools" that does the heavy lifting here, is Pega's open sourced CDH utilities library mentioned earlier. It can be installed directly from GitHub following the instructions in https://github.com/pegasystems/cdh-datascientist-tools.
Importing the PMML file into Pega
Once you have the PMML files, they can be imported from Prediction Studio. Create a model (or update one)
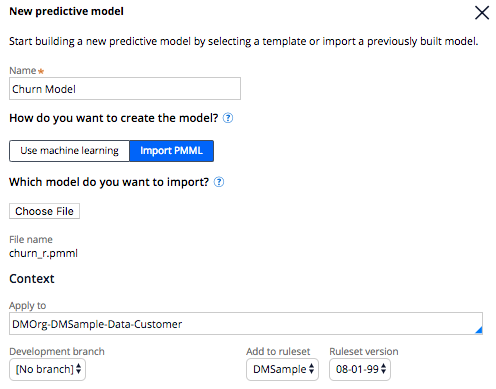
give it a name, indicate "import PMML", select the PMML file and specify the context (class) that this model is for - in our examples DMOrg-DMSample-Data-Customer. If you work directly in DMSample you may want to create a ruleset for yourself:
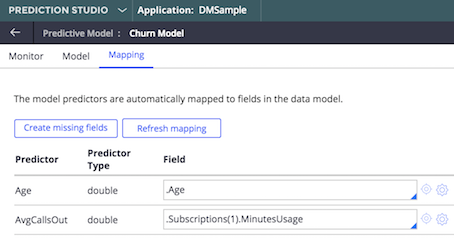
You may be prompted for some additional meta for monitoring purposes. When the import of the PMML file is done, review the mapping of the input fields. In our example Age is available in the DMSample Customer class, but AvgCallsOut is not. You could map it the same way DMSample maps it (see the Predict Churn model), passing in the usage number from the first subscription.
Using the model in Pega
Now that the model has been imported, you can use it on it's own or use it like any other model component in your strategies. Use the "Run" facility to run the model and interactively provide the inputs.

The model predicts a higher probability of churn for younger people with a high usage pattern. Makes sense.
Out of the box, DMSample uses a PMML model for Risk and a Pega model for Churn. You could replace the Churn model by one of the PMML models, for example.
Summary
The sklearn2pmml and r2pmml libraries are powerful tools to export Python scikit-learn and R models to PMML. Both support the inclusion of preprocessing steps in the PMML. This is even more important for the Python models than for the R models as the scikit-learn classifiers generally assume numeric inputs, while many of the R classifiers can work with symbolics directly.
For classifiers that are not included (yet) in the umbrella packages there often are specialized libraries available to convert to PMML, like for XGBoost and LightGBM.

Hi
I was trying to replicate this and see the results. But have got stuck somewhere. Please help.
Steps done:
1. created the PMML file
2. have given the path of that file to be imported. Specified the apply to field.
3. there is no option to select the ruleset and the ruleset version.
Please suggest!
Thanks!